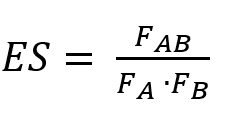KMAP¶
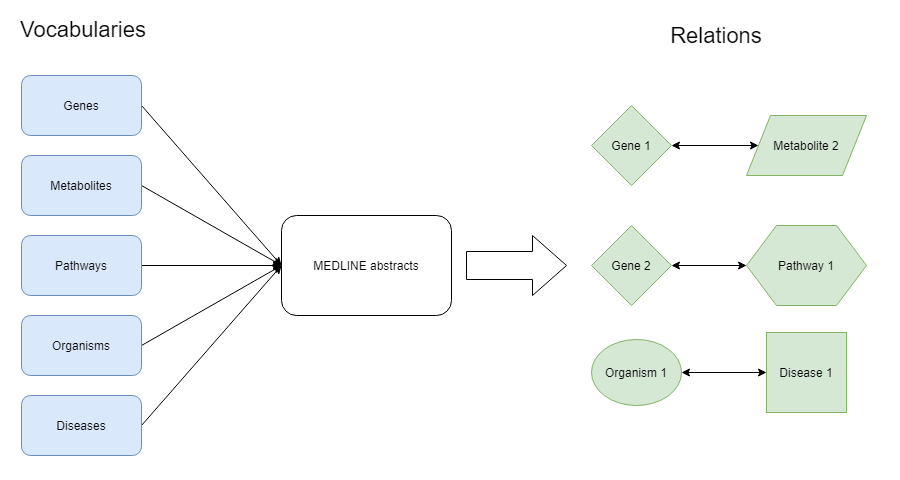
KMAP is a knowledge database generated in a semi-automated fashion from the scientific literature, in this case, millions of MEDLINE abstracts. A schematic overview of the construction of KMAP is shown below. We have constructed a large database with biological keywords, organized in vocabularies describing genes, pathways, diseases, organisms etc. which we refer to as concepts. We used these concept vocabularies to search the entire MEDLINE database of ~35 million abstracts. Based on the results we constructed relations between concepts that are statistically associated with each other in the literature.
Concept¶
The keywords in the vocabularies are referred to as biological concepts, and are organized in vocabularies. Each concept always has the following properties:
An identifier that uniquely identifies the concept. An identifier is constructed with a prefix followed by a number. From the prefix, the vocabulary can be deduced.
A name that is the most common name for the biological concept.
Next to that, a concept can have synonyms and additional identifiers (xrefs) that link the concept to other databases.
Relation¶
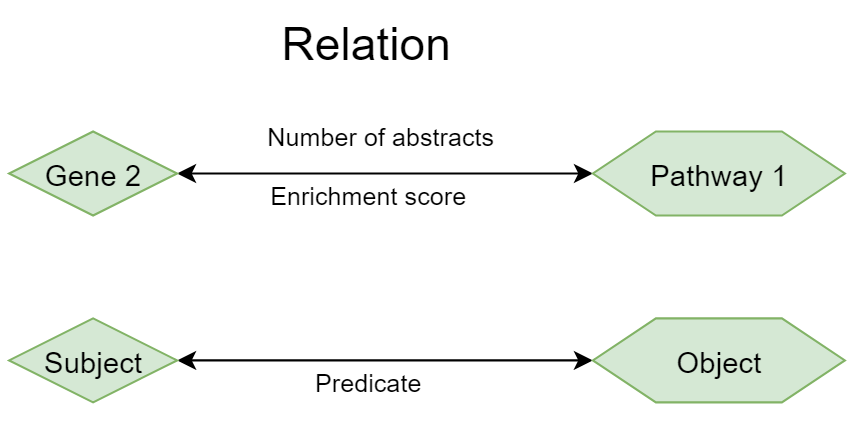
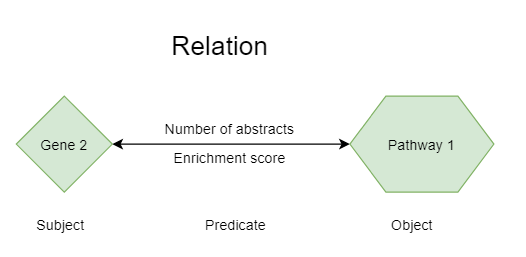
A relation is defined between two concepts, when they occur together in an abstract or in a sentence. A single abstract in which the two concepts occur is sufficient to define a relation. Of course, when the concepts occur together in more abstracts, the relation is probably stronger. A relation always has a Subject (the first concept) and an Object (the second concept) and a type of relation (Predicate). In the case that two concepts occur in an abstract, the Predicate is “Co-occurrence” and the relationship is reciprocal. If we analyze a relationship on sentence level, we may be able to more specifically infer the nature and the direction of the relation.
To characterize the strength of the relation, we can look at 3 numbers.
Number of abstracts. This is the number of abstracts in which both concepts co-occur with each other.
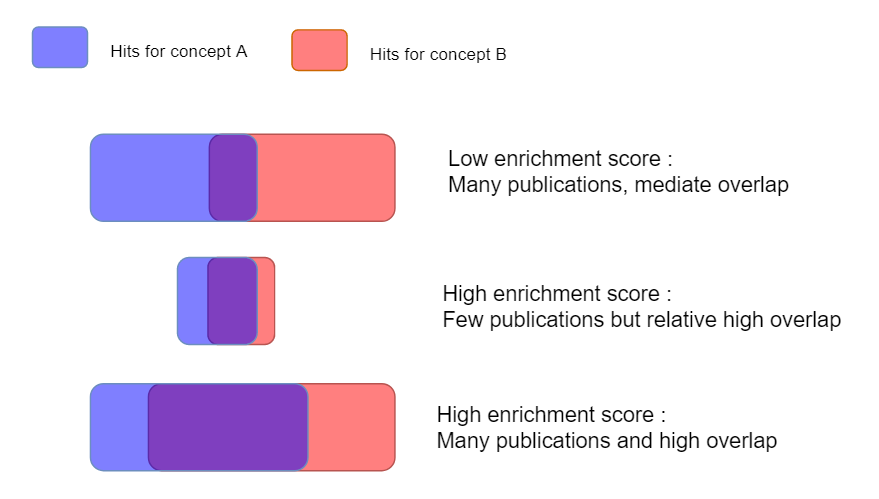
The enrichment score (ES). This score is based on the Mutual Information (MI) score and corrects for publication bias in concepts. For example, the term “cancer” has a lot of hits in PubMed and consequently each concept would have automatically a strong hit with cancer if we would score on abstracts alone, but this would not necessarily imply a strong, specific connection (see also figure below).
In practice the ES tends to be very high for relations in which both concepts only have a few hits in MEDLINE whereas the number of abstracts is biased towards well-known concepts with a lot of abstracts. The local MI is the product of the ES and number of abstracts and is in practice a good trade-of. For further theory on scoring word associations, see the thesis of Stefan Evert.
The formula that is used to calculate the Enrichment Score (ES) is
In which FAB is the number of abstracts in which both concepts co-occur and FA and FB are the number of abstracts in which the concepts occur separately.
Concept Sets¶
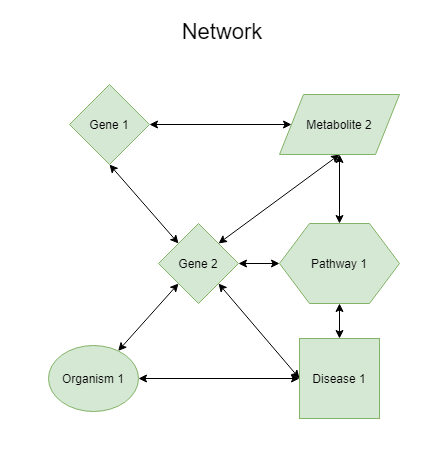
A concept set can be regarded as a set of concepts that have one or more mutual relations which each other. A natural representation of such a concept set is a network in which all pairwise relations are shown. Typically concept sets can be :
a set of regulated genes derived from a gene expression experiment.
a set of diseases that is tied to a specific SNP or region of the chromosome.
a set of molecular pathways with their corresponding genes.
a set of micro-organisms obtained from a metagenomics experiment.
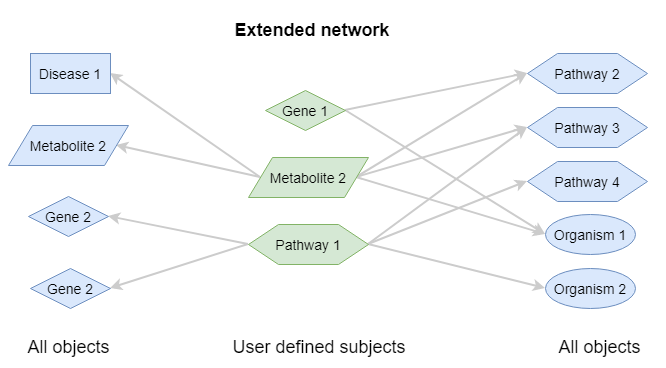
These concept sets can be used to find interesting network patterns, but can also be used to find additional terms that are related to this set as whole. For example, a new concept that would have a connection with all of the members of the concept set would be interesting (such as Pathway 2 in the figure).
Reference sets¶
All of the information is deduced from text. Currently KMAP works mostly on abstracts, although full text analysis is also possible. Abstracts sets can be used for the deduction of relations between concepts, but can also be used to quickly gather relevant literature around a subject. So, apart from searching with concepts, the API can also be approached with a starting set of PMIDS that are obtained from another source or application.
Access to the API¶
Input¶
The input required for the API is a URL indicating the type of function that is required and a list of one or more parameters. Most of the time these parameters will be a number of identifiers for abstracts, concepts or vocabularies and parameters that modify the results by e.g., setting a limit on the number of results that are returned, ordering the results or the exact format of the returned results
Parameter type |
Format |
Description |
|---|---|---|
single identifier input |
A string representing a single identifier |
An identifier is used to uniquely pinpoint a single vocabulary, concept or abstract. |
identifier_list input |
A comma separated list of single identifiers |
Lists are sued to indicate multiple identifiers, e.g. when working with sets of concepts and sets of abstracts. |
format of the output |
A number or string controlling the output. |
These parameters are used to limit the results to a maximum, order the results or formatting the results in a specific way. |
Calls & Results¶
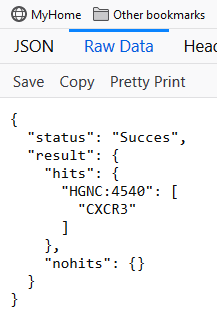
Each call the servers produces a result in JSON format. The standard response is a dictionary with 2 entries. For example the call concept/search/?terms=CXCR3&apikey=MY_VALID_APIKEY returns the following JSON output
The status field gives an indication of the outcome of the run. currently this has the following values
Success. The execution of the call was OK
Input Error. Some required arguments are missing or not correctly formatted.
Internal server Error. Something went wrong on the server side. If this problem persists in multiple calls, please contact us.
The result field returns the actual results. In the case that the call was OK, there will be the actual results. In the case of an Input Error, a relevant help message is displayed. In case of an Internal server error, the message “The server could not process your request for an unknown reason.” is displayed.
Access keys¶
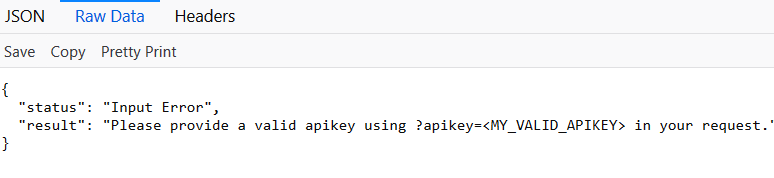
You need an APIKEY to get access to the API methods. The APIKEY This is a code that needs to be used in the scripts and send to the server each time an API request is done. Please contact us at *kmine@tenwise.nl* for getting a trial API-key. If you do not supply an APIKEY in your request the server will give an error and ask you to supply an APIKEY.
Without supplying an apikey, the server returns an Input Error
**concept/search/?terms=CXCR3 **
GET vs. POST requests¶
All methods work both as a GET and POST method. In the GET methods all parameters and their values are supplied in a URL format that can be directly executed in the browser. In a POST request, the parameters and their values are supplied in the body of a form. You can use GET request for simple calls to the server, but if you have large concept sets or abstracts sets to send to the server, you should use POST, because URLS can only have a maximum length of characters and may be dependent on your browser. When running a POST request, you are required to include an additional cookie in your request as a security protection. Please see the R and Python examples on how to do this.
Methods¶
The following tables give an overview of the methods that are available, with a short description. In general, each call needs to be executed with a number of parameters. The exact parameters that need to be used in each call are detailed in the documentation for each call. Parameters that are required are in bold. Parameters that are optional can be omitted and the default value is shown between [].
Concept methods¶
These methods are preceded by the /concept/ statement in the URL and return data and relations for individual concepts.
Method name |
Function |
Input |
Output |
|---|---|---|---|
search |
Searches the database for concepts that match your keywords |
A set of keywords or external identifiers. |
The concept identifiers that match to your keywords. |
hits |
Returns the MEDLINE abstracts in which the concept occurs. |
A concept identifier. |
A list of PMIDs. |
relations |
Returns related literature concepts. |
A concept identifier. |
A set of related concepts and the statistical scores. |
concept/search¶
Searches a concept on basis of the supplied strings and returns a dictionary with the matching concepts. The strings can be ordinary keywords but also valid identifiers to other databases such as entrez gene ids, disease ontology terms, Ensembl gene ids etc.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key as a string. |
|
terms |
A string with comma separated search terms. |
|
wildcard |
[false] true |
If true, adds a wildcard to the string. So e.g. searching for CXCR with a wildcard will return all chemokine receptors such as CXCR3, CXCR5, CXCR6 etc. |
Returns (Example)
Value |
Description |
|---|---|
hits |
A dictionary with as keys the concept identifiers that were found. As values a list of one or more terms from the search string that matched to this identifier. |
nohits |
A dictionary with as keys the terms for which no hits were found. |
concept/hits¶
This method returns the references in which a concept is found. A hit is characterized by the PMID of the reference, the number of hits in the reference and by the string_hit, i.e., the string to which the concept matches in the abstract. This can be informative in case you want to see whether the hit was with the preferred name of the concept or with one if it’s synonyms. In case of multiple hits in the reference, only the string_hit for the first encountered term is given.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_id |
The identifier of the concept for which the results should be retrieved. |
|
retmax |
[100] |
The maximum number of hits to retrieve. |
ordering |
[pmid] hitnr |
The ordering is done on PMID, so effectively returning the most recent references. If ordering is set to hitnr, the references with the most hits of hits concepts are returned. |
Returns
Value |
Description |
|---|---|
hits |
A list of dictionaries with as key-value pairs the PMID, the hitnr and the string_hit. |
concept/relations¶
This method returns the relations for a given concept. A relation is defined as a co-occurrence of two concepts in a reference (see example below), identified by a subject and object, connected by a predicate. See here for a more extended description of what we mean by a relation.
The relations can be ranked on basis of the
overlap
the enrichment score (escore)
local_mi, which is the product of the overlap and the enrichment score and as such balances the influence of both parameters.
By default, all relations for a concept are returned. By setting the vocab_ids parameter, the relations can be restricted to certain vocabularies only.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_id |
The identifier of the concept for which the results should be retrieved. |
|
vocab_ids |
string |
A comma separated list of vocabulary_ids for which the relations need to be retrieved. If not supplied, all relations are returned. |
ordering |
[local_mi] overlap escore |
The way the ordering should be done. |
retmax |
[50] |
The maximum number of relations to return after ordering |
Returns (Example)
Value |
Description |
|---|---|
hits |
A list of dictionaries with as key-value pairs the abs_id(=PMID), the hitnr and the string_hit. |
Relation methods¶
These methods are preceded by the /relation/ statement in the URL and return details on a relation between concepts. Currently there is only one relation method, but this will be extended in the future.
Method name |
Function |
Input |
Output |
|---|---|---|---|
evidence/ |
Returns the references for a relation. |
Two concept identifiers. |
A set of PMIDs in which the concepts co-occur. |
relation/evidence¶
Returns all the PMIDs for a relation, along with the number of hits for both concepts in each PMID.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
subject |
The concept_id for the subject of the relation. |
|
object |
string |
The concept_id for the object of the relation. If not supplied, defaults to the subject, so effectively retrieving the references in which the subject occurs with itself. This is normally not what you want, but the server does not throw an error if you do. |
ordering |
[total_hits] pmid |
The default is to order on the number of total hits, i.e. the references in which the concepts have a lot of hits. If ordering is done on PMID, it returns in practice the most recent references. |
retmax |
[100] |
The maximum number of relations to return after ordering. |
Returns (Example)
Value |
Description |
|---|---|
evidence |
A list of dictionaries with as with the subject_id, the object_id, the pmid and the hits for each of the concepts. |
Reference Set methods¶
These methods work on a set of MEDLINE abstracts, which are referred to by their identifiers, the PMIDs. PMIDs are numbers, ranging from 1 to ~35.000.000 that uniquely identify a PubMed reference.
Method name |
Function |
Input |
Output |
|---|---|---|---|
citations/ |
Returns citation information. |
A list of PMIDS |
Citation information for each PMID. |
concepts/ |
Returns the concepts that are found in the references. |
A list of PMIDS |
For each reference the concepts and associated hits. |
markup/ |
Highlights the concepts in the actual references and individual sentences |
A list of PMIDS and a list of concepts to highlight. |
Strings that can be used for display as highlighted HTML. |
free_search/ |
Returns PMIDs based on a free text string |
A string |
A list of PMIDs. |
class_scores/ |
Returns a classification score for a set of PMIDS |
A list of PMIDS |
A set of classification scores for the PMIDS. |
refset/citations¶
This method returns basic annotation information for a set of PMIDs. This citation information includes amongst other the title, abstract, journal, publication date. This should be sufficient to reconstruct the most important citation information. If you are interested in more details, e.g. author affiliation, ISNN number of the journal etc., you can retrieve the data directly from PubMed using their Entrez system. The fields that are returned by this function are as follows:
Field |
Description |
|---|---|
pmid |
The PMID, a number of 2 or more digits |
title |
The reference title as text |
abstract |
The abstract as text |
journal_title |
The abbreviated journal title |
volume |
The volume |
issue |
The issue |
page |
The page(s) |
authors |
“;” separated full names of the authors e.g. ‘Marie Krogh Nielsen; Torben Lykke Sørensen’ |
meshterms |
“;” separated MESH terms e.g. ‘Aged;Case-Control Studies;Chemokine CCL5;’ Can be an empty string. |
chemicalterms |
“;” separated chemical terms. Can be an empty string. |
pubmed_date |
Date in the format YYYY-MM-DD, e.g. ‘2020-04-24’ |
NB. Currently we have placed a cap of 1000 on the number of references that can be retrieved in a single query. If you need to retrieve more than 1000 references, please use this call in a loop, with no more 1 call per ~2 seconds.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
pmids |
A string with a number of “,” separated PMIDs. |
Returns (Example)
Value |
Description |
|---|---|
citations |
A list of dictionaries with citation information for the abstracts, as described above. |
refset/concepts¶
This method returns the concepts for a set of PMIDs. This can thus be regarded as the reverse as the function to get the PMIDs for a set of concepts. The use case for this is that you have somehow retrieved a set of PMIDs e.g. from a PubMed query, or from and Endnote library or a reference list from a paper, and you want to look up the concepts that are referred to in these abstracts. It can also be used to prioritize a large list of PMIDs to only those ones that contain a lot of concepts of your interest.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
pmids |
A string with a number of “,” separated PMIDs. |
|
vocab_ids |
string |
A string with an optional parameter specifying the vocabularies from which the concepts should be returned. This can be used to limit the results of the call. If this parameter is not specified, all concepts are returned. |
Returns (Example)
Value |
Description |
|---|---|
pmid2concept |
A dictionary with PMIDs as keys and a list of concept_ids as values |
refset/markup¶
This function takes a set of PMIDs and concept_ids and returns highlighted versions of the abstracts or the sentences in the abstracts. Only the abstracts and sentences in which at least 1 concept is found are returned. When you set the highlight parameter to False, the non-highlighted version of the text is returned. This can make sense if you want to do your own analysis on the sentences in which a co-occurrence is found. The combination of markup_mode=’abstract’, highlight =False does not make a lot of sense, since you would be basically returning the plain abstracts, which can easier be obtained via another way. Currently the highlighting can be done in two ways
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
A string |
Your API key |
pmids |
string |
A string with a number of “,” separated PMIDs. |
concept_ids |
string |
A string with concept_ids. |
retmode |
[highlight] plain_text |
Whether to return highlighted text. |
Returns (Example)
Value |
Description |
|---|---|
markup |
For each PMID a dictionary with the a title and abstract and sentences, in which the concepts are enclosed in <span style=\”background-color:#ADD8E6;\”> </span> tags if retmode is “highlight”. |
refset/free_search¶
All methods in this API are geared towards retrieving evidence for relations that have been precomputed based on the relations between the concepts. However, sometimes it is necessary to start with PMIDs that are not based on a concept, but on a free text search on the entire MEDLINE library. Therefore serves this particular method. This method returns PMIDs for a free text query. Individual terms in the query that are not quoted are translated into an “OR” query. To combine terms with an explicit “AND”, you need to also use this also in the query. See the table below for examples (please not that an APIKEY needs also to be provided). The method is based on an index file that is being created using the lucene indexer from Apache.
Query |
Hits |
Remarks |
|---|---|---|
?terms=cxcr3 |
3549 |
Simple query |
?terms=cxcr? |
31900 |
Wild card at the last position, retrieves all CXCR abstracts |
?terms=cxcr3%20ccr6 |
4886 |
Implicit OR |
?terms=cxcr3%20AND%20ccr6 |
372 |
Explicit AND |
?terms=cxcr3%20OR%20ccr6 |
4886 |
Explicit OR |
?terms=”cholesterol%20metabolism” |
6486 |
Quoted phrase |
?terms=cholesterol%20AND%20metabolism |
57545 |
Explicit AND |
?terms=(cholesterol%20OR%20steroid)%20AND%20metabolism |
73555 |
OR and AND combination |
The following arguments are required:
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
A string |
Your API key |
terms |
string |
A search string. |
retmax |
[50] |
The maximum number of PMIDs to return. The PMIDs are reverse sorted on number, effectively yielding the most recent PMIDs first. |
Returns (Example)
Value |
Description |
|---|---|
query |
The query that was executed. |
pmids |
A list of PMIDs. |
hitnr |
The number of hits that were found. This is not the same as the number of PMIDs that are returned, this is governed by the retmax parameter. |
refset/class_scores¶
With this function, you can retrieve scores for a s et of abstracts for a number of classification models. These classification models have been developed on basis of a RandomForest model that was trained on a number of article classes. Currently we score for the following article classes.
Clinical : Articles related to clinical trials
Variant : Article related to genetic variation
Mouse : Articles related to experimental mouse models
Rat : Articles related to experimental mouse models
Microbiome : Articles related to microbiome experiments
3R : Articles related to experiments aimed at replacement of animal models
All models scores range from 0 (not related) to 1 (very much related). We only return articles with a score higher than 0.5.
The following arguments are required:
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
A string |
Your API key |
pmids |
string |
A string with a number of “,” separated PMIDs. A maximum of 2000 PMIDS is allowed at this moment. |
Returns (Example)
Value |
Description |
|---|---|
class_scores |
A list of class_score results. Each result contains the score for one or more classes. If a class score is below 0.5 it is not included in the output. |
Concept Set methods¶
These methods are preceded by the /conceptset/ statement in the URL and return data and relations for sets of concepts. When running this on a lot of concepts, you may need to use the POST request in order to avoid overloading the URL.
Method name |
Function |
Input |
Output |
|---|---|---|---|
annotation/ |
Returns details for concepts, such as synonym, xrefs etc. |
A set of one or more concept identifiers. |
A list of annotated concepts. |
relations/ |
Retrieves related concepts for each concept in the set. |
A set of concept identifiers. |
A set of new, related concepts. |
enrichment/ |
Retrieves related concepts for the set as a whole using an enrichment calculation. |
A set of concept identifiers. |
An enrichment table with related concepts and the p-value. |
evidence/ |
Retrieves most relevant references for a set of concepts. |
A set of concept identifiers. |
A set of PMIDs along with a relevance score. |
conceptset/annotation¶
With this function, detailed on information on the concepts can be obtained. This includes information on the name and synonyms but also references to other external databases. See example below:
Not all concepts have the same level of detail. For genes, much more information is available then for certain metabolites. The only annotation tags that are always available are the ‘name’ and ‘concept_id’ tag.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_ids |
A string with a number of “,” separated values. |
Returns (Example)
Value |
Description |
|---|---|
annotation |
A list of dictionaries (one for each concept identifier) with tag-value pairs for the annotation fields. |
conceptset/relations¶
This function connects the concepts with a new set of concepts. The parameters in the call can be used to control how the relations should be created. By setting these parameters you can build the following networks.
Extended network¶
This is built by connecting all concept ids to all target concepts from the databases that match to one or more of these concept ids. Optionally, the target concepts can be limited to only a certain vocabulary.
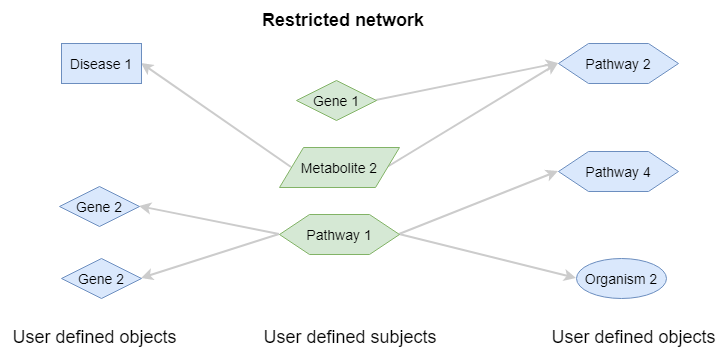
Restricted network¶
This is similar to the extended network, but now only relations are shown between user specified target concepts. This is an interesting method if you have a set of genes and you are interested in the links to certain prespecified diseases or pathways of interest.
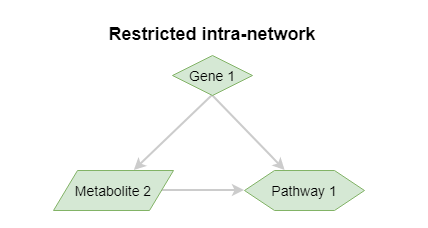
Restricted intra-network¶
A special form of the restricted network is the intra-network, which can be obtained by specifying the objects with the same identifiers as the subject identifiers. You then only get links between the original concepts in the data set.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_ids_subject |
A string with a number of “,” separated values. |
|
concept_ids_target |
string |
A string with a number of “,” separated values that will be used to limit the output to only show relations with the specified targets. |
vocab_ids |
string |
A string with a number of “,” separated values specifying the vocabularies for which the targets should be retrieved. |
Returns (Example)
Value |
Description |
|---|---|
connectivity |
For each node in the network a specification of how many other nodes it is connected to. |
relations |
A list of dictionaries, one for each relation. The keys of the dictionaries describe the relation, and is explained here. |
concept_set/enrichment¶
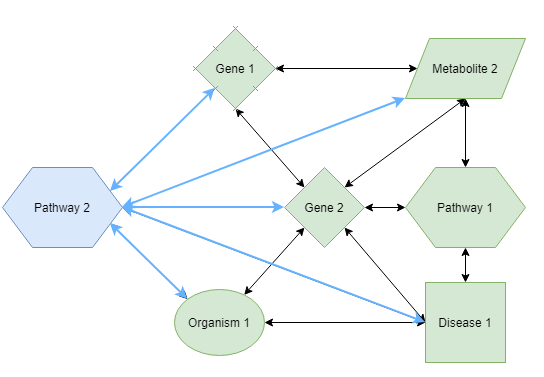
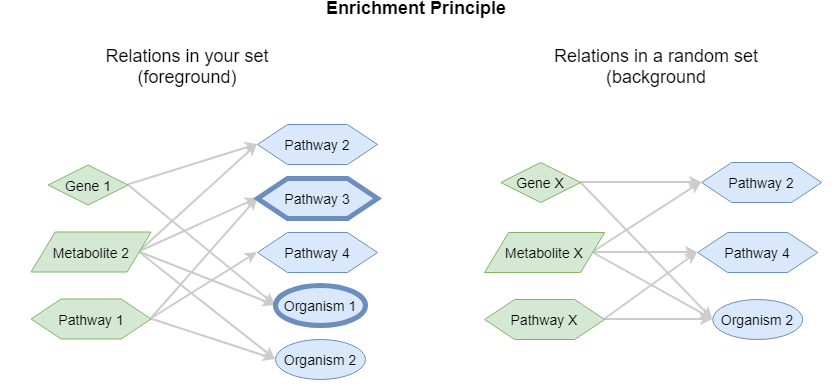
This function returns the enriched concepts for the set of concept_ids. This function is related to the relations function, but with the difference that a statistical analysis on the results is performed. This statistical analysis ensures that only concepts are retained that have a stronger connection to your input set compared to a random background set. This analysis has been described as Gene Set Enrichment Analysis (GSEA), but the principle can be performed on any set of concepts. The principle is demonstrated in the following figure:
The left network shows the relations with your concepts of interest (foreground set). The right panel shows the relations in a random set of the same composition, i.e. the same number of concepts from each vocabulary type (background set). This shows that Pathway 3 and Organism 1 are specifically connected to the foreground set whereas Pathway 2, Pathway 3 and Organism 1 are connected to both sets and thus less specific for the foreground set. The specificity / relatedness for the foreground set can be calculated by a so-called contingency table on which a Fisher Exact test can be applied. The p-value is an indication of how specific the new concepts are for your concept set (i.e. the foreground set).
The outcome of this method is very much dependent on what exactly you choose as a background set. This is often dependent on your research question and sometimes difficult to create. If you do not specify a background set in the method, we will match your foreground set with respect to the number of concepts and distribution across vocabularies. However, since this is a random process, you may expect changes in the results when rerunning the method, especially with smaller sets.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_ids |
A string with a number of “,” separated values. |
|
background_ids |
string |
An optional string with background_ids |
Returns (Example)
Value |
Description |
|---|---|
enrichment |
A dictionary with the enriched objects. Each element has the slots as explained below. |
background_set |
A set of concept ids that formed the background set. This is either equal to the one supplied by the user, or the automatically generated one. Especially in the last case, saving this background set and using it in future reruns of your call will get you reproducible results. |
The results in the enrichment dictionary are as follows.
id: The concept id of the associated concept.
pval: The P-val for the association as determined.
hitsfg: The number of hits with the foreground set.
nohits: The number of no-hits with the foreground set.
hitsbg: The number of hits with the background set.
nohitsbg: The number of no-hits with the background set.
enrichment: The fraction of hits with the foreground set compared to the hits in the background set.
ratio: The natural log of the enrichment.
targets: The concept ids in the foreground set for which a relation was found.
conceptset/evidence¶
This function returns the PMIDs that have the highest coverage of concepts in your set. For example, if you have a set of 5 concepts, and in a single abstract, 4 of those 5 concepts occur, this abstract gets a score of 0.8. Because this method potentially can return a lot of results there is a cap of 1000 PMIDs returned per function call. By default, the ordering is done on the score, meaning that the best covered references are returned. Ordering on the PMID yields the most recent references.
A special case can occur if you have a large concept set and want to retrieve PMIDS that at least always contain a single of multiple prespecified concept_ids. These can be specified with the anchor parameter. Returned PMIDS are then guaranteed to have at least one of those concept_ids.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_ids |
A string with a number of “,” separated values. |
|
hitlimit |
[2] |
An integer specifying the number of concepts that should at least be present in the abstracts. Setting this to 1 also returns abstracts in which only one concept occurs, which may be interesting for sparsely connected concept sets. |
ordering |
[score] pmid |
The ordering of the results, either on most relevant (score) or most recent (pmid). |
anchor |
None |
A set of concept_ids. These can be used to specify that all returned PMIDs should at least contain a hit to all the ids specified by the anchor ids. |
retmax |
[50] |
The maximum number of PMIDs to return after applying the hitlimit and ordering. |
Returns (Example)
Value |
Description |
|---|---|
evidence |
A list of dictionaries with the PMID, the score and the number of hits. |
conceptset/hits¶
This function returns all the hits for a set of concepts. This means that for each concept, all the PMIDs in which the concept has a hit, are returned. This is analogous the concept/hits method, but now repeated for all concepts in the set. Because this method potentially returns a lot of data, the number of concepts in the set that can be used for this call is limited to 500.
Parameter |
Value [default] |
Description |
|---|---|---|
apikey |
Your API key |
|
concept_ids |
A string with a number of “,” separated values. |
Returns (Example)
Value |
Description |
|---|---|
hits |
A dictionary with the PMIDs as keys and a list of concepts from the set as values |
Vocabularies¶
The following vocabularies are currently in use.
ID |
Name (prefix of the IDS) |
Description |
Number of concepts |
|---|---|---|---|
ONT0001 |
HGNC Genes (HGNC) |
A curated online repository of HGNC-approved gene nomenclature. |
42357 |
ONT0003 |
Human Disease (TWDIS) |
A vocabulary for human disease representation. |
17615 |
ONT1004 |
Tool Compounds (TOOLC) |
A set of pharmaceutical tool compounds. |
16061 |
ONT0005 |
Organisms (TAX) |
A set of terms covering a large subset of the NCBI taxonomy database. |
519716 |
ONT0006 |
Pathways (PATH) |
A set of terms covering a large set of biological pathways. |
2798 |
ONT0008 |
Cell types (TWCELL) |
A set of terms covering a large set of cell types. |
117 |
ONT1005 |
Bacterial Genes (BACG) |
A set of terms describing bacterial gene symbols. |
10857 |
ONT1006 |
Metabolites (TWMET) |
A set of terms describing metabolites. |
1214 |
ONT5003 |
GenesComp (TWHGNC) |
A set of additional terms describing gene complexes. |
4 |
ONT1007 |
Workflow terms (TWRWO) |
A set of terms describing workflow terms. |
86 |
ONT1008 |
Food terms (TWFOOD) |
A set of terms describing food items. |
2875 |
Workflows¶
This section details a number of workflows that can be used to address common problems. A workflow is defined here as a series of API calls, where the results of one API call are used by the subsequent API calls. The shuttling of data between the API calls can be done within Python, R or on the command line with shell scripts.
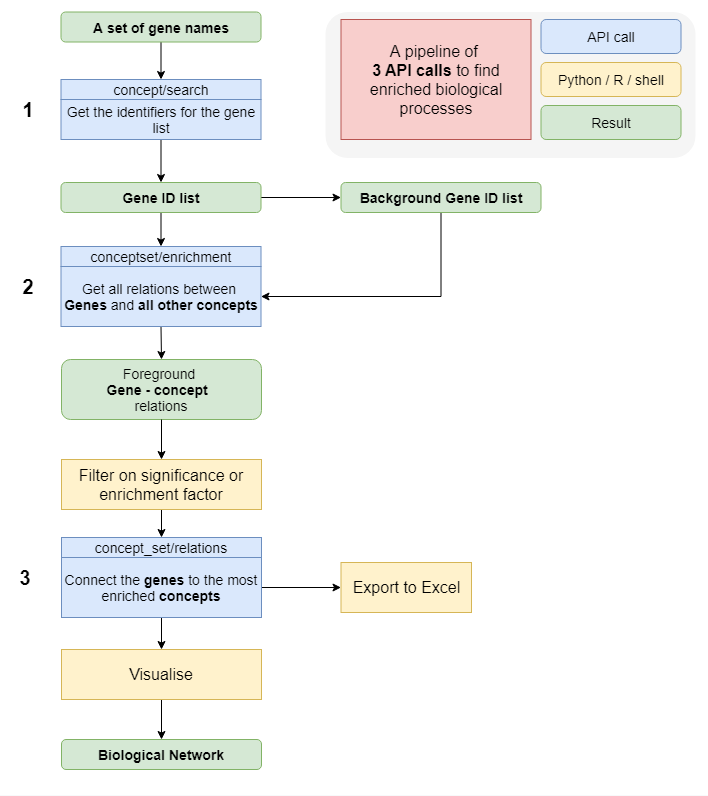
Gene set enrichment analysis¶
This workflow is designed to start with a list of genes and to perform a Gene Set Enrichment analysis.
An explanation of the steps:
1: A keyword search is carried out to obtain the correct identifiers for the for the gene names.
2: An enrichment call is done to get the concepts that are preferentially connected to the gene set of interest. Optionally a background set can be applied.
3: The most enriched concepts are retrieved from the results and a network visualization is created. Alternatively, the output can be exported to Excel.
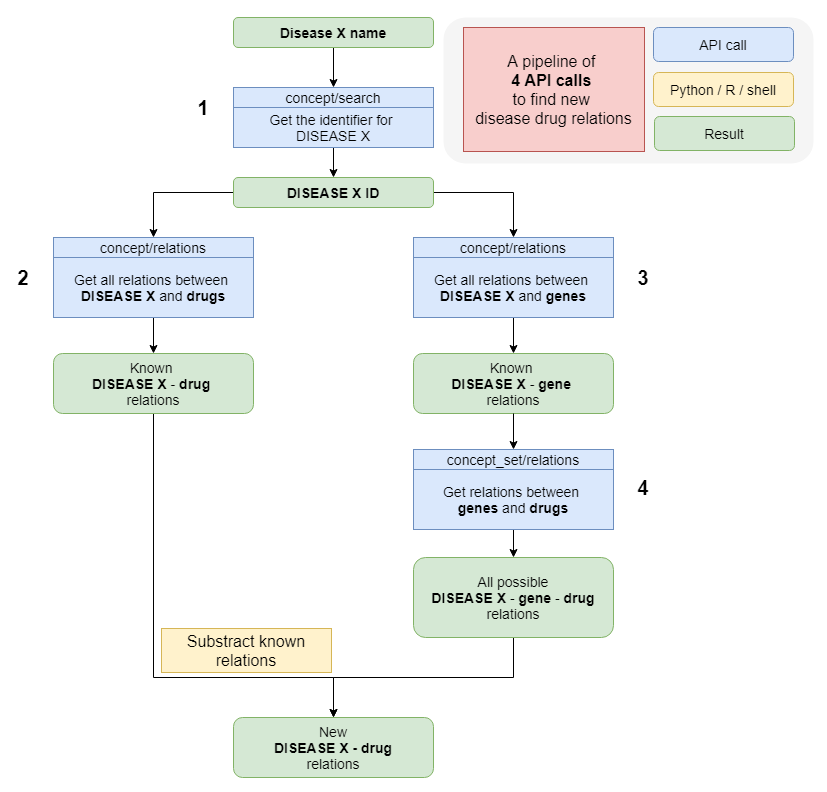
Drug repurposing¶
This workflow is designed to discover new relations between concepts based on a set of shared intermediates. This is also called the ABC approach. An example of the application of an ABC approach using this API interface can be found here.
An explanation of the steps:
1: A keyword search is carried out to obtain the correct identifier for the disease term.
2: All known relations with drugs are obtained by executing the relations call, limiting the results to the drug vocabulary. This yields known A-C relations.
3: All known relations with genes are obtained via the relations call, limiting the results to only genes. This establishes the A-B relation in the ABC principle.
4: The retrieved genes are treated as a concept set, and the entire concept set is matched to new drugs. This establishes all B-C relations in the ABC principle.
The A-B and B-C data sets can now be connected via overlapping B-nodes. This yields all potential A-B-C relations. To find truly new relations, subtract known A-C relations from the found A-B-C relations.